Lecture Details[]
Rod Devenish; Week 5 MED1011; Biochemistry
Lecture Content[]
Difference in protein production pattern from different cells is differential transcription, many mechanisms may result in selective transcription (as a result of a signalling event), transcriptional control is a major control point for the regulation of gene expression in eukaryotic cells. Proteins regulate transcription initiation, and level of transcription is achieved by protein combination. Many but not all of these proteins bind directly to DNA.
Multiple regulatory sequences are the sites of protein binding, promotor sequence determines the site of transcription initiation and binding or RNA polymerase II and transcripton apparatus. Enhancers are located up to 50kb up/downstream of a promoter within an intron, contain binding sites for proteins that stimulate transcription. Repressors turn off transcription, compete with activators for binding sites on DNA, mask activator activity or have a direct negative effect on transcription complex. Binding sites for these proteins are generally <20bp. Proteins that directly bind DNA have partially complementary features. Many contacts can join together for added adhesion.
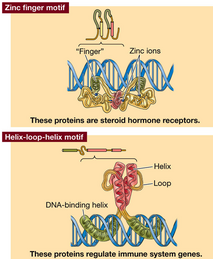
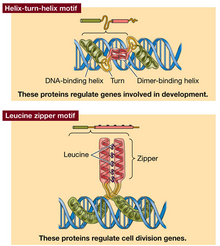
Four motifs for protein-DNA interaction: helix turn helix (regulate genes of development); leucine zipper (regulate cell division); zinc finger (steroid hormone receptors); helix loop helix (regulate immune system genes). Some regulatory proteins are dimers to increase options, if monomers have different binding specificity then different number of regulatory sequences can be bound. Some zipper proteins dimerise with a partner unable to bind DNA so block DNA binding.
Non-DNA binding proteins recognise a surface from DNA binding proteins (protein complex). Changes in nucleotide sequence of a binding site changes 3D structure of bound proteins and influences further binding. Can recruit activators or repressors. Activity can be controlled by protein synthesis, ligand binding, phosphorylation, addition of another subunit, unmasking (removal of inhibitor), stimulation of nuclear entry (inhibitor removed to allow entry into nucleus), release from membrane.
miRNA (19-25 nucleotides) base pairs with mRNA targets, expressed in tissue specific fashion. siRNA regulates transposons, viral infection. piRNA is essential for developing germ cells.
DNA that has been packaged by nucleosomes is transcriptionally silenced, inactive chromatin is silenced by proteins that render it inaccessible, for other regions silencing is reversible. Chromatin loops isolate the active chromatin. The base of the loop is an insulaitng element, protein binds. Barrier sequences stop the spread of gene activation by tethering chromosomes to fixed sites, binding proteins to groups of nucleosomes, recruiting histone modifying enzymes which erase marks required for activation.
Regulation of chromatin assembly/structure requires ATP dependent chromatin remodelling (from energy of ATP hydrolysis) and histone modifications. Chromatin remodelling complexes bring about a change in structure (histone acetyltransferase loosens nucleosome to DNA attachment). Proteins are recruited (remodelling proteins, disaggregating the nucleosome), some changes affect nucleosomes. Transcription complex then binds and begins. Affinity for DNA and chromatin associated proteins is controlled by modification of H2A, H2B, H3, H4. Histones have N terminal tails which extend from surface of nucleosome.
Histones can be modified by acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, glycosylation, which produce patterns of modification and mediate protein-protein interactions. Acetylation of all 4 core histones occurs on all conserved lysines. Is reversible. Correlation between hyperacetylation and activation and hypoacetylation and inactivation/silencing. Methylation controls gene expression, differentiation and development, preservation of chromosomal integrity, X inactivation, involved in brain function and the immune system. Occurs at 5' position of cytosine in the sequence, does not affect base pairing. Methylation pattern is stable but not irreversible and is inherited in replication. It is stamped on DNA without changing its sequence (epigenetic). Methylated C's are found in clusters or CG islands within or near the promoter of 60% of genes. Binding of proteins to clustered methylated C's results in transcription repression (histone deacetylase binds to protein/methyl compound). Linkage between DNA methylation and transcription repression.
Genomic imprinting is a result of methylation, determined by whether it is inherited by the mother or father. Pattern of methylation is known as imprinting. Physical basis is that one (mother or father gene) is silenced by methylation. In germ line cells, all genes are demethylated. In gametes, sex specific methylation pattern is established (de novo methylation). Methylation patterns are reprogrammed in early embryo (demethylated and remethylated). Somatic cells have heavy methylation, trophoblast derived cells (placenta, yolk sac) are less methylated, primordial germ cells are unmethylated. Disease occurs if active parental copy of gene is mutated, mutation occurs in genes controlling imprinting, genes that encode activators/repressors are imprinted leading to altered target gene expression. Hypomethylation leads to chromosome instability, activation of endogenous parasitic sequences, loss of imprinting. Hypermethylation leads to inactivation of key cell pathways in DNA repair.
In cloning, demethylation/de novo methylation does not occur in donor nucleus cells before they are in germ line state, likely explains why cloning is ineffective. In females one X chromosome is methylated, expresses 1 gene, Xist which transcribes RNA that is non-coding, remains in nucleus, deactivates X chromosome. Also called a Barr body.